Tracking Tiny Organisms Using Big Data
Editor’s note: Each semester, students in the Geospatial Analytics Ph.D. program can apply for a Geospatial Analytics Travel Award that supports research travel or presentations at conferences. The following is a guest post by travel award winner Megan Coffer as part of the Student Travel series.
Cyanobacteria, commonly referred to as blue-green algae, are only micrometers in size––a thousand times smaller than a millimeter. Thousands of them can fit in a single drop of water. And yet, we can see these microscopic organisms using satellites orbiting hundreds of kilometers above Earth’s surface. When enough cyanobacteria are present in a lake, they actually change the way light is absorbed and reflected, and these changes can be tracked with multispectral sensors. This is where my research is focused: using satellite imagery to track cyanobacterial blooms in lakes across the United States.
Cyanobacterial blooms can negatively affect human health, the environment and the economy. Human exposure can lead to anything from an irritating rash to flu-like symptoms. Fish suffer from decreased oxygen levels and aquatic plants struggle to get sunlight under large blooms. Economic impacts include medical treatment, loss of tourism, monitoring costs and commercial fishery impacts.
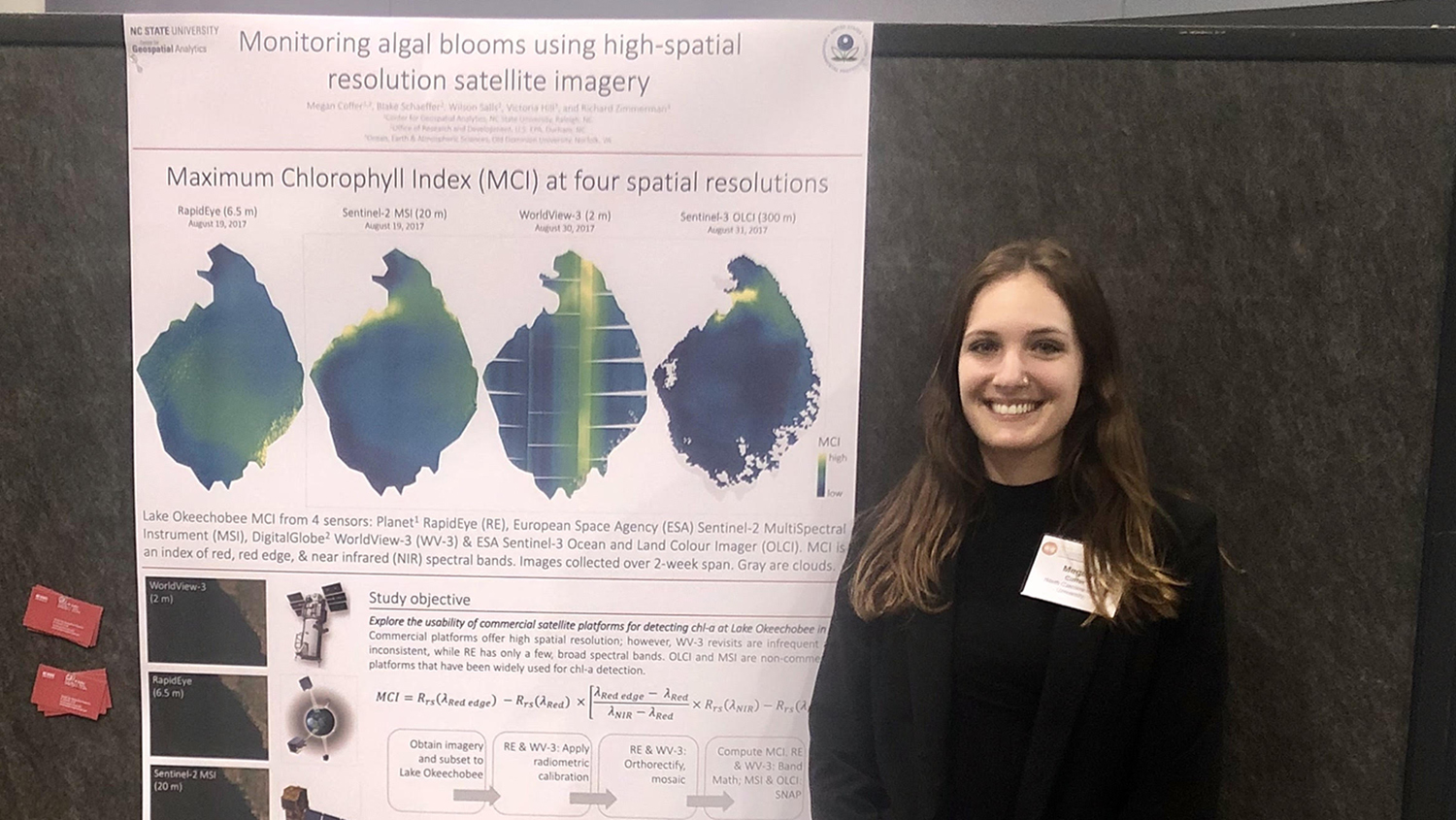
Traditionally, we use low-cost satellites that offer coarse spatial resolution, usually with a 300-m pixel size. This means that the smallest features the sensor can identify are about as big as 13 soccer fields. Recently, I have begun exploring the possibility of detecting cyanobacteria using commercial satellite platforms. These satellites offer a spatial resolution up to 150 times finer than the coarse-resolution sensors currently in use. Although commercial satellites also have their drawbacks (their data can be expensive, and they can’t be relied upon to fly over the same location to collect repeat data at consistent times), they offer an intriguing option for addressing spatial limitations inherent in our traditional approach.
Last November, I received a travel grant from the Center for Geospatial Analytics to attend the Coastal and Estuarine Research Federation (CERF) meeting in Mobile, Alabama, where I presented my findings regarding the usability of two commercial satellites for cyanobacteria detection. DigitalGlobe’s WorldView-3 satellite offers a spatial resolution of 2 m and Planet Lab’s RapidEye satellite constellation offers a spatial resolution of 6.5 m. This improvement in spatial resolution allows patterns within cyanobacterial blooms to be distinguished, which can reveal important characteristics of large-scale cyanobacterial blooms not evident when studying smaller populations. Additionally, higher-resolution imagery enhances the ability to see shorelines, where recreation typically occurs and where drinking water intakes tend to be located.
A primary outcome of this research has been the development of a standardized workflow for processing data from commercial platforms. Few studies have used data from these platforms because of their high price tag, and so there is little guidance for how to use their data most effectively. The workflow I developed takes imagery from basic products, as delivered from each company, and turns them into analysis-ready products usable for a variety of scientific applications. Our research group believe this to be the first processing protocol of its kind, and will make it available for public use upon publication of our results.
At CERF, I presented results from a study using commercial satellite imagery to compute the abundance of cyanobacteria at the water’s surface at Lake Okeechobee in Florida. I found that both WorldView-3 and RapidEye demonstrate promise for detecting cyanobacterial blooms at a fine spatial scale, as compared to traditional approaches. My research suggests that traditional satellites are still the best option for monitoring large areas for potentially harmful blooms, but commercial platforms can provide unprecedented detail to examine blooms on an individual basis.